
Here are the point estimates and uncertainties from the glm fit and stan_glm fit, which we see are nearly identical: The documentation for stan_glm has more information about these controls as well as other topics related to GLM estimation. The stan_glm function has many other optional arguments that allow for more user control over the way estimation is performed. We also provided a seed so that we have the option to deterministically reproduce these results at any time.

The chains argument controls how many Markov chains are executed, the cores argument controls the number of cores utilized by the computer when fitting the model. We’ve also specified some optional arguments.
#For counter 1 to x estimating pbasic code
The poisson family function defaults to using the log link, but to write code readable to someone not familiar with the defaults we should be explicit and use family = poisson(link = "log"). The formula, data, family, and offset arguments to stan_glm can be specified in exactly the same way as for glm. Library(rstanarm) data(roaches) # Rescale roaches $roach1 <- roaches $roach1 / 100 # Estimate original model glm1 <- glm(y ~ roach1 + treatment + senior, offset = log(exposure2), data = roaches, family = poisson) # Estimate Bayesian version with stan_glm stan_glm1 <- stan_glm(y ~ roach1 + treatment + senior, offset = log(exposure2), data = roaches, family = poisson, prior = normal( 0, 2.5), prior_intercept = normal( 0, 5), seed = 12345) \[\tfrac\) to the linear predictor \(\eta_i\) and it can be specified using the offset argument to stan_glm. If the outcome for a single observation \(y\) is assumed to follow a Poisson distribution, the likelihood for one observation can be written as a conditionally Poisson PMF This vignette focuses on Step 1 for Poisson and negative binomial regression models using the stan_glm function. Steps 3 and 4 are covered in more depth by the vignette entitled “How to Use the rstanarm Package”.
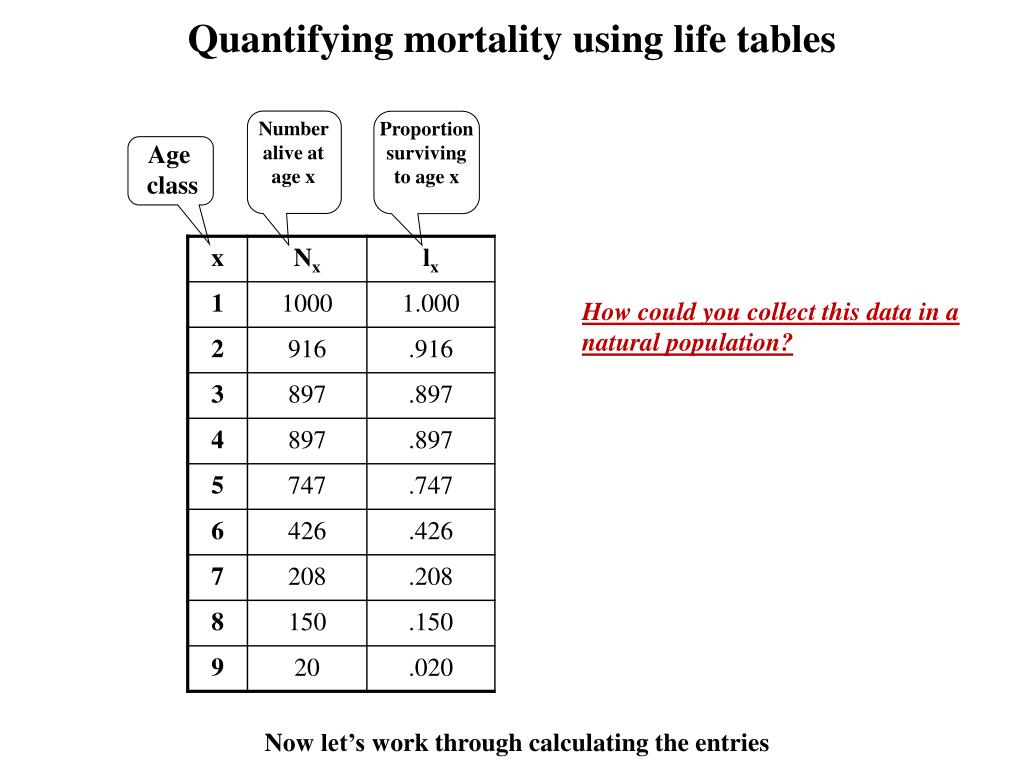
This joint distribution is proportional to a posterior distribution of the unknowns conditional on the observed data Specify a joint distribution for the outcome(s) and all the unknowns, which typically takes the form of a marginal prior distribution for the unknowns multiplied by a likelihood for the outcome(s) conditional on the unknowns.The four steps of a Bayesian analysis are
#For counter 1 to x estimating pbasic how to
This vignette explains how to estimate generalized linear models (GLMs) for count data using the stan_glm function in the rstanarm package.


 0 kommentar(er)
0 kommentar(er)
